-
If you are citizen of an European Union member nation, you may not use this service unless you are at least 16 years old.
-
You already know Dokkio is an AI-powered assistant to organize & manage your digital files & messages. Very soon, Dokkio will support Outlook as well as One Drive. Check it out today!
| |
Nucleic Acids and Proteins
Page history
last edited
by Darrell Sharp 12 years, 5 months ago
 |


|
 |


|
|
| DNA |
Nucleotide |
Nucleotides ==> DNA |
Amino Acids, Peptide Bonding, and Protein |
|
|
|
|
|
|
7.5 Proteins
Amino Acid Song - shows the 20 amino acids (you don't have to know them)
|
Homework Questions
PowerPoint
Click4BIology
|
|
|
|
7.5.1
Explain the four levels of protein structure, indicating the significance of each level.
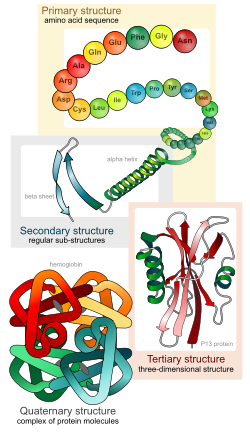
|
Watch this video: Protein Structure
The four levels of protein structure:
1. Primary: the sequence of amino acids
2. Secondary: alpha helices and beta pleated sheets - held together by hydrogen bonds
3. Tertiary: overall strucutre of one polypeptide created by interactions between side groups (R groups)
4. Quaternary: large proteins made of multiple polypeptides
Another video showing several different models of proteins.
|
|
|
| 7.5.2
Outline the difference between fibrous and globular proteins, with reference to two examples of each protein type.
|
Two groups of proteins:
1. Fibrous proteins
- long, fibrous shape
- mostly alpha helices or beta pleated sheets
- insoluble - don't dissolve in the water of the cell
- function as structural components - building blocks
Examples
- keratin: hair, skin, nails
- collagen: in skin, bones, teeth, blood vessels,
2. Globular proteins
- soluble in water - dissolve in the cytoplasm of the cell
- diverse structure - compact but not symmetrical
- diverse functions: transport, storage, signals, catalysts (enzymes)
- (includes all proteins that are not structural (fibrous)
Examples
- hemoglobin: transports oxygen
- catalase: breaks down peroxide
|
|
|
| 7.5.3
Explain the significance of polar and non-polar amino acids.
|
Polar and Nonpolar Amino Acids
Some R groups are polar, and some are nonpolar
The whole protein can have a polar or nonpolar characteristic
Or the protein can have polar and nonpolar regions
Polar likes polar. They interact.
Polar hate nonpolar. They don't interact.
Vocabulary
hydrophilic: "water loving" polar characteristic
hydrophobic: "water fearing" nonpolar characteristic
Look at this diagram of the cell membrane with a "channel protein" (its function is to transport the red circles through the membrane).

http://www.click4biology.info/c4b/7/pro7.5.htm#three
Nonpolar regions (pink) interact with the hydrophobic (nonpolar) region of the plasma membrane.
Polar regions (green) interact with the hydrophilic (polar region) of the plasma membrane.
|
|
|
| 7.5.4
State four functions of proteins, giving a named example of each.
|
Function - Example
Catalyst (enzyme) - Catalase: breaks down peroxide in cells
Transport - Hemoglobin: transports oxygen in the blood
Hormone - Insulin: signals cells to absorb glucose from the blood
Structure - Keratin: in skin, hair, nails
Defence - Antibodies: kill bacteria that enter the body
|
|
|
|
|
|
|
|
7.1 DNA structure

http://web.nmsu.edu/~snsm/classes/chem435/Lab4/
|
Homework Questions
Click4Biology
|
|
|
|
7.1.1
Describe the structure of DNA, including the antiparallel strands, 3’–5’ linkages and hydrogen bonding between purines and pyrimidines.
|
DNA is a double helix consisting of two strands of polynucleotides. The two strands are held together by hydrogen bonds between the complementary bases in the interior of the double helix. The exterior of the double helix, or "backbone" of the strands, is made of the phosphate and deoxyribose subunits of the nucleotides.
Antiparallel Strands
The two strands of the double helix are oriented in opposite directions.
The strand is referred to as having a 5' to 3' direction.
- The carbon atoms of deoxyribose are numbered 1 to 5
- The phosphate is attached to the 5' carbon. The base is attached to the 1' carbon
- The phosphate of the next nucleotide is attached to the 3' carbon of the preceding nucleotide
- The bond goes from the 3' to the next phosphate.
Each strand has the opposite 5' to 3' direction = antiparallel.
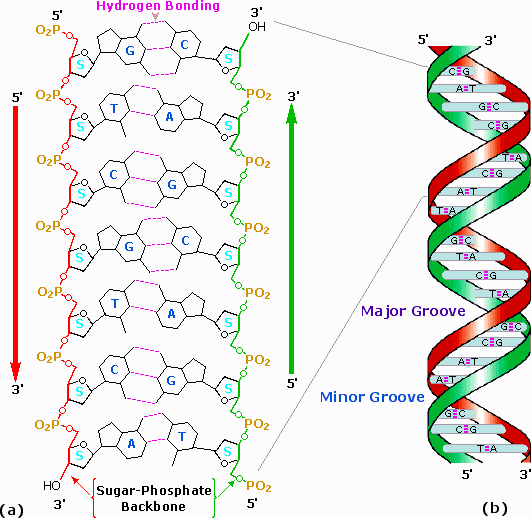
http://www2.chemistry.msu.edu/faculty/reusch/VirtTxtJml/nucacids.htm
Complementary Base Pairing
- Purines = Adenine and Guanine (big)
- Pyrimidines = Thymine and Cytosine (small)
- Complementary bases are always one big base (purine) and one small base (pyrimidine).
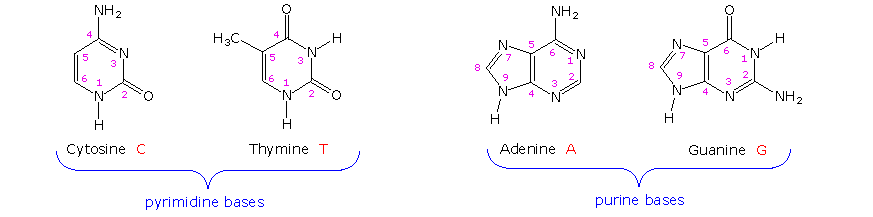
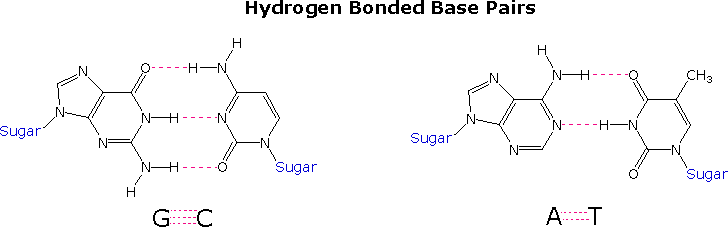
http://www2.chemistry.msu.edu/faculty/reusch/VirtTxtJml/nucacids.htm
|
|
|
| 7.1.2
Outline the structure of nucleosomes.

Supercoiling Video
|
Human DNA is approximately 2 meters long!
How does it fit into the nucleus of a cell?
Answer: supercoiling
Supercoiling
DNA is wrappped around histone proteins to create a nucleosome.
The histone proteins are like the spool that string is wrapped around.
This makes it more condensed.

All the DNA and histone proteins are called chromatin.
The chromatin is coiled and coiled and coiled....to produce a chromosome.

http://www.chromatintoronto.ca/projects.html
|
|
|
| 7.1.3
State that nucleosomes help to supercoil chromosomes and help to regulate transcription.
|
Nucleosomes control how DNA is used.
Besides helping to fit all the DNA into the nucleus, nucleosomes help to regulate transcription.
Transcription is "reading" the genes to control the cell or organism.
- When DNA is wrapped tightly around the histones, it cannot be "read"
- When DNA is wrapped loosely around teh histones, it can be "read"
|
|
|
| 7.1.4
Distinguish between unique or single-copy genes and highly repetitive sequences in nuclear DNA.
|
Not all DNA has important information.
Unique or single copy genes have specific information for controlling the cell or organism
Highly repetitive sequences do not contain information.
Most of the DNA is actually highly repetitive sequences, also called noncoding.
Genes are a very small percentage of the total DNA.
|
|
|
|
7.1.5
State that eukaryotic genes can contain exons and introns
|
Within the genes, there is also extra noncoding sequences.
- Exons are parts of genes that are expressed to control the cell or organism.
- Introns are parts of genes that are removed as the DNA information is copied.
The noncoding DNA between genes and the introns within genes do have functions.
- The highly repetitive regions protect the genes from random mutations. The long regions of "junk" DNA reduce the probability that a mutation will happen in an important region, a gene.
- The introns within the genes allow one gene to produce several products, depending on which introns are removed. The "editing" of DNA information also makes a step where the process can be controlled - faster, slower, or stopped.
|
|
|
|
|
|
|
|
7.6 Enzymes
Apoptosis simulation
|
Homework Questions
PowerPoint
Click4Biology
Lab Report Template
Enzymatic Activity Lab
|
|
|
|
7.6.1
State that metabolic pathways consist of chains and cycles of enzyme-catalysed reactions.
|
Metabolism: all the chemical reactions in a cell or organism.
- Catabolic: reactions that break down big molecules into small molecules.
- Anabolic: reactions that build big molecules from small molecules.
Metabolic pathways can be chains or cycles.
Each stage has its own enzyme
Chain pathways go from substrate to product.
From Click4Biology:

- Enzyme (1) is specific to substrate 1. This is changed to product 1.
-
Enzyme (2) is specific to product 1 which becomes the substrate and converted to product 2.
-
Enzyme (3) is specific to products which becomes the substrate and converted t o product 3.
-
Product 3 is called the 'End product'.
Example: Glycolysis
Cyclic pathways begin with an intermediate molecule that bonds with the substrate.
- The intermediate molecule is re-formed in the pathway
- So its "cyclic"
From Click4Biology:

-
Enzyme (1) combines the regenerated 'intermediate 4' with the initial substrate to catalyse the production of intermediate 1.
-
Enzyme (2) is specific to intermediate 1 and converts intermediate 1 to intermediate 2
-
Enzyme (3) is specific to intermediate 2 and catalyses it conversion to product and intermediate 3.
-
Enzyme (4) is specific to intermediate 3 and catalyses its conversion to intermediate 4.
-
The difference is the regeneration of the intermediate, in this case intermediate 4.
Examples: Krebs cycle and Calvin cycle.
|
|
|
|
Chain Pathway: Glycolysis
 
Cycle Pathway: Krebs Cycle

|
|
|
| 7.6.2
Describe the induced-fit model.
|
Induced Fit Model
The "induced-fit" model improves the "lock and key" model.
- Some enzymes are not specific to one substrate (like thelock and key).
- They catalyze several similar substrates.
- The enzyme changes shape as the substrate enters the active site.
- This is the "induced-fit" - the substrate causes the active site to change shape.
From Click4Biology:

|
|
|
| 7.6.3
Explain that enzymes lower the activation energy of the chemical reactions that they catalyse.
|
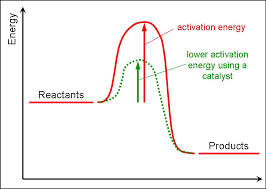
Enzymes Lower Activation Energy
Activation energy is the energy needed to start a chenical reaction.
Enzymes speed up chemical reactions by lowering the activation energy.
They are easier to start, so more reactions happen in less time.
|
|
|
| 7.6.4
Explain the difference between competitive and non-competitive inhibition, with reference to one example of each.

|
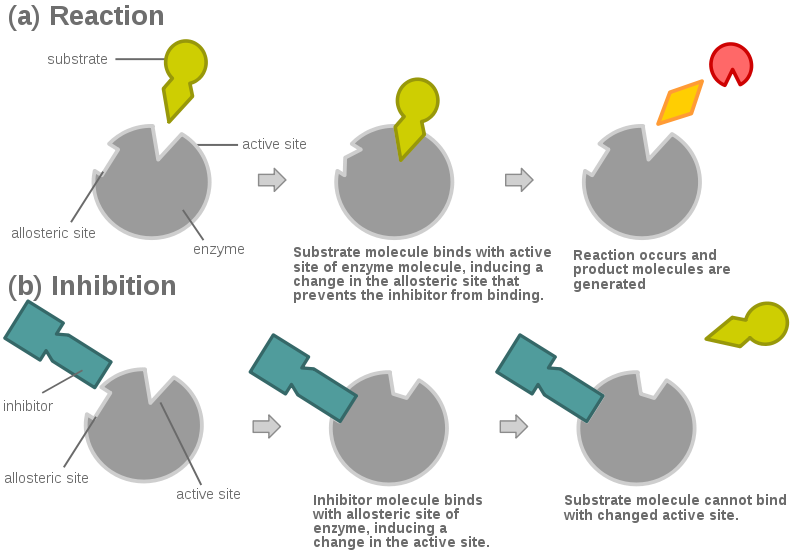
Inhibitor: a molecule that slows or stops an enzyme's activity.
Competitive Inhibition
- The inhibitor molecule is similar to the substrate.
- It competes with the substrate for the active site of the enzyme.
- It can bind to the active site.
- This slows or stops the reaction.

Non-Competitive Inhibition
- The inhibitor is different than the substrate.
- It cannot bind to the active site.
- It binds with another region of the enzyme - the allosteric site.
- This changes the shape of the enzyme and its active site.
- So the substrate cannot bind to the active site.
- So the reaction is slowed or stopped.
|
|
|
| 7.6.5
Explain the control of metabolic pathways by end-product inhibition, including the role of allosteric sites.
|
End-Product Inhibition
The product of the metabolic pathway is the inhibitor.
- End-product inhibition can be competitive or non-competitive.
- The enzyme has an allosteric site where the non-competitive inhibitor binds.
- This kind of inhibition regulates metabolic pathways to control the amount of product
Video
Video - Thanks, Jessica!
|
|
|
|
|
|
|
|
7.2 DNA replication
|
Homework Questions
Click4Biology
|
|
|
|
7.2.1
State that DNA replication occurs in a direction. direction.
7.2.2
Explain the process of DNA replication in prokaryotes, including the role of enzymes (helicase, DNA polymerase, RNA primase and DNA ligase), Okazaki fragments and deoxynucleoside triphosphates.
|

http://genmed.yolasite.com/fundamentals-of-genetics.php
Look at the diagram above as you read the notes below.
The dark blue strands are the original DNA (called "Parental" in the diagram).
Notice the two strands are antiparallel. What does this mean?
Helicase unzips the DNA.
Replication begins with the enzyme RNA primase adding complementary RNA nucleotides called an RNA primer.
DNA polymerase III starts adding DNA nucleotides to the RNA primers.
Look in the little box to see the red primer at the beginning of the leading strand.
Notice the new DNA strands are light blue (called the leading strand and lagging strand).
See that the leading strand is replicated continuously from its primer.
See that the lagging strand is replicated discontinuously (in short pieces) from many primers.
DNA polymerase III only adds nucleotides to the 3' end of a nucleotide.
It builds in a 5' to 3' direction.
The lagging strand is replicated in short pieces because it's antiparallel to the leading strand.
The short pieces are called Okazaki fragments.
DNA polymerase III builds an Okazaki fragment then releases the lagging strand and reattaches nearer to the replication fork.
The two parts of DNA polymerase III stay together.
Another enzyme called DNA polymerase I replaces the RNA primers with DNA nucleotides.
The enzyme ligase joins the Okazaki fragments together.
DNA Replication video
_______________________________________________________________
Just to make it more complicated, the lagging strand is actually looped around so that DNA polymerase III can work on both strands at the same time.
This diagram is actually more accurate:

http://www.macroevolution.net/biology-dictionary-rare.html#.Tp_PbZsr2so
DNA Replication - looped lagging strand
DNA Replication Video 2 (more realistic - my favorite)
_______________________________________________________________
Nucleoside triphosphates
- The free nucleotides floating around actually have three phosphate groups.
- They are called nucleoside triphosphates.
- During replication, when the free nucleotide is bonded to the growing strand, the two extra phosphates are removed.
- This provides energy for the reaction.
- In the diagram below, notice the nucleoside triphosphate at the bottom left being bonded to the growing strand in a 5' to 3' direction.
- The two phosphates are released.

http://iws.collin.edu/biopage/faculty/mcculloch/1406/outlines/chapter%2015/chap15.html
|
|
|
|
7.2.3
State that DNA replication is initiated at many points in eukaryotic chromosomes.
|
Notice in the diagram below that there are two replication forks moving in opposite directions.

http://patticarothersonline.org/Archived%20Biology%20Items/dna_replication_diagram_of_leadi.htm
This is called a replication bubble.
DNA replication is started at many sites along the molecule.
This makes DNA replication happen quickly for a extremely long molecule.

|
|
|
|
|
|
|
|
7.3 Transcription
|
Homework Questions
Click4Biology
|
|
|
|
7.3.1
State that transcription is carried out in a  direction. direction.
|
In transcription, the RNA molecule is built in the 5' to 3' direction.

http://dna-rna.net/wp-content/uploads/2011/08/rna-transcription2.jpg
|
|
|
|
7.3.2
Distinguish between the sense and antisense strands of DNA.
|
In DNA, only one strand is transcribed into RNA.
The strand of DNA that is used as a template for RNA is called the antisense strand.
The other strand of DNA is called the sense strand and it is identical to the RNA except for T's instead of U's
("it makes sense")

https://buffonescience9.wikispaces.com/UNIT+4b-Gene+Expression+
|
|
|
|
7.3.3
Explain the process of transcription in prokaryotes, including the role of the promoter region, RNA polymerase, nucleoside triphosphates and the terminator.

|
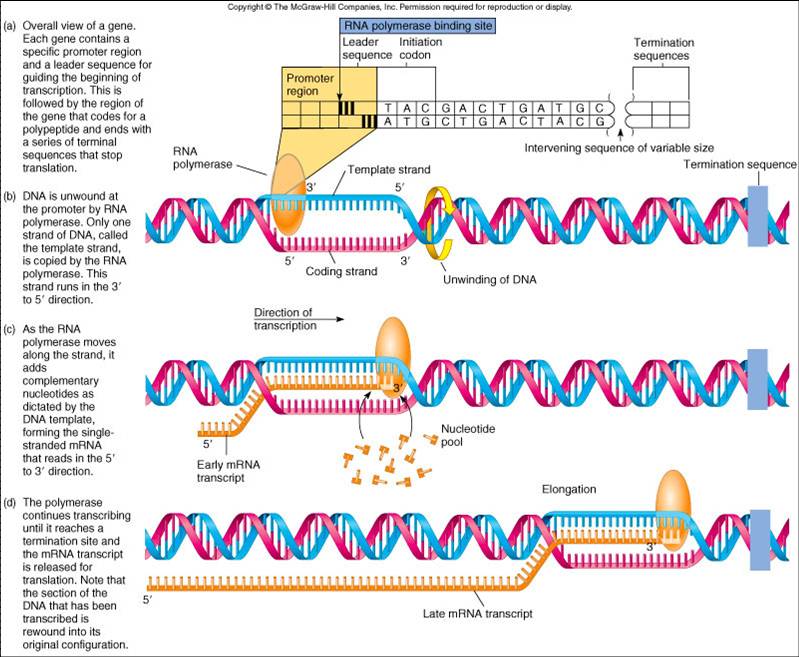
Transcription
1. Initiation
- Helicase unwinds the DNA.
- RNA polymerase binds to the promoter region of the DNA.
- The promoter region has a pattern of nucletides that allow RNA polymerase to know the antisense strand, the start of a gene, and the correct direction.
- Nucleoside triphosphates with ribose sugars and the bases A, U, C, and G hydrogen bond to the DNA.
2. Elongation
- RNA polymerase makes covalent bonds between the nucleoside triphosphates which release energy for the reaction.
- The RNA strand is built in the 5' to 3' direction.
3. Termination
- At the end of a gene is a region of the DNA called the terminator.
- The terminator is a pattern of bases that causes the RNA polymerase to release from the DNA.
- The completed RNA is also released from the DNA.
- DNA rewinds into its double helix form.
Transcription video
- the video shows more detail than required by IB.
- note the number of proteins that are necessary for transcription.
|
|
|
|
7.3.4
State that eukaryotic RNA needs the removal of introns to form mature mRNA.
|
Transcription produces RNA.
- in prokaryotic cells, the mRNA is ready for translation.
- in eukaryotic cells, mRNA processing occurs before translation.
mRNA Processing
RNA produced by transcription is composed of regions called introns and exons.
- The introns are removed during mRNA processing.
- The exons are spliced together during mRNA processing.
- mRNA that is used for translation is composed of only exons.
- "Exons are expressed"

|
|
|
|
|
|
|
|
7.4 Translation
|
Homework Questions
Click4Biology
|
|
|
|
7.4.1
Explain that each tRNA molecule is recognized by a tRNA-activating enzyme that binds a specific amino acid to the tRNA, using ATP for energy.

|
There are 64 possible codons for mRNA.
So there are 64 possible anticodons for tRNA.
There are 20 amino acids coded for by the 64 codons.
The correct amino acid needs to matched to the correct tRNA anticodon.
Enzymes called tRNA-activating enzymes bind with specific tRNA molecules and specific amino acids and bond them togther.
This requires energy from ATP.
The specificity of these enzymes is important for accurate translation.
The animation shows the amino acid valine (Val) being added to a tRNA.

Note: that tRNA molecules are reused after they bring amino acids to the ribosome.
|
|
|
|
7.4.2
Outline the structure of ribosomes, including protein and RNA composition, large and small subunits, three tRNA binding sites and mRNA binding sites.
|
Ribosome Structure
- composed of a mix of protein and rRNA
- small subunit - holds the mRNA
- large subunit - holds the tRNAs and bonds the amino acids together
- A site - where the tRNA with an amino acid enters the large subunit ("amino acid")
- P site - where the tRNA with the amino acid chain is held ("polypeptide")
- E site - where the tRNA without an amino acid exits the ribosome ("exit")
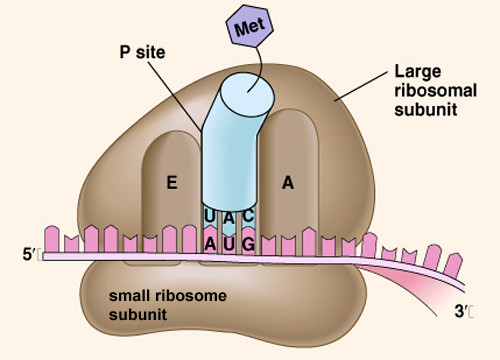
Note: at the beginning of translation, the first tRNA occupies the P site.
|
|
|
|
7.4.3
State that translation consists of initiation, elongation, translocation and termination.
7.4.4
State that translation occurs in a  direction. direction.
7.4.6
Explain the process of translation, including ribosomes, polysomes, start codons and stop codons.
|
Translation
1. Initiation
- An activated tRNA with the correct anticodon binds to an mRNA at the start codon (AUG).
- A small subunit of a ribosome binds to the mRNA at the start codon.
- A large subunit of a ribosome binds with the small subunit.
- The first tRNA is in the P site of the ribosome.
 (Initiation) (Initiation)
2. Elongation
- An activated tRNA with the correct anticodon to match the second codon of the mRNA enters the A site.
- The ribosome catalyzes the formation of a peptide bond between the adjacent amino acids.
- The ribosome also catalyzes the breaking of the bond between the first tRNA and its amino acid.
3. Translocation
- The ribosome moves in a 5' to 3' direction along the mRNA.
- The first tRNA, now without its amino acid, moves into the E site and released.
- The second tRNA, with the growing polypeptide, moves into the P site.
- A new tRNA enters the A site and matchs its anticodon to the mRNA codon.
- Elongation of the polypeptide happens simultaneously with translocation of the ribosome.
 (elongation and translocation) (elongation and translocation)
4. Termination
- When the ribosome reaches a stop codon of the mRNA, a protein called a release factor enters the A site.
- The release factor causes the bond between the tRNA in the P site and the polypeptide to break.
- The polypeptide is released and folds into its tertiary structure.
- The ribosomal subunits separate and release the mRNA.
 (termination) (termination)
Animations from Pearson http://www.phschool.com/science/biology_place/biocoach/translation/init.html
Polysomes
One mRNA molecule may have many ribosomes attached to it at one time.
They follow each other like a train and make multiple copies of the polypeptide.

Translation video
Ribosome (from wikipedia)

|
|
|
|
7.4.7
State that free ribosomes synthesize proteins for use primarily within the cell, and that bound ribosomes synthesize proteins primarily for secretion or for lysosomes.
|
Ribosomes have two locations in eukaryotic cells.
- free ribosomes float in the cytoplasm. They produce proteins for use in the cell.
- bound ribosomes are attached to the endoplasmic reticulum. They produce proteins for secretion from the cell or for use in lysosomes.
|
|
|
|
7.4.5
Draw and label a diagram showing the structure of a peptide bond between two amino acids.
|

|
|
|
|
|
|
|
|
|
|
|
|
|
Nucleic Acids and Proteins
|
|
Tip: To turn text into a link, highlight the text, then click on a page or file from the list above.
|
|
|
|
|
Comments (0)
You don't have permission to comment on this page.